Atomic force microscopy, a tactile-based probe technique, provides a three-dimensional nanoscale image of a material by gliding a needle-like arm across the material's surface. The core of AFM imaging workhorse is a cantilever with a sharp tip that deflects as it encounters undulations across a surface. Due to a minimum force required for imaging, conventional AFM cantilevers can deform or even tear apart living cells and other biological materials. While scientists have made strides in reducing this minimum force by making smaller cantilevers, the force is still too great to image cells with high resolution. Indeed, for imaging objects smaller than the diffraction limit of light—that is, nanometer dimensions—this approach hits a roadblock as the instrument can no longer sense minute forces.
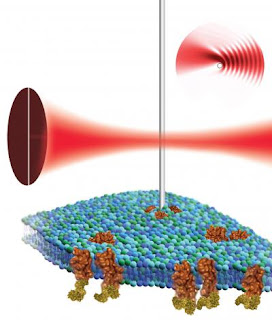
Caption: By placing a nanowire cantilever in the focus of a laser beam and detecting the resulting light pattern, scientists at the Molecular Foundry could use atomic force microscopy to non-destructively image the surface of a biological cell (green and blue structure) and the proteins (shown in brown) associated with it.
Credit: Illustration by Flavio Robles, Berkeley Lab Public Affairs. Usage Restrictions: None.

Caption: Babak Sanii (left) and Paul Ashby with Berkeley Lab's Molecular Foundry, have designed a nanowire-based imaging tool to study living cells and other soft materials in their natural, liquid environment.
Credit: Photo by Roy Kaltschmidt, Berkeley Lab Public Affairs. Usage Restrictions: None. | Now, however, scientists with the Molecular Foundry, a U.S. Department of Energy User Facility located at Berkeley Lab, have developed nano-sized cantilevers whose gentle touch could help discern the workings of living cells and other soft materials in their natural, liquid environment. Used in combination with a revolutionary detection mechanism, this new imaging tool is sensitive enough to investigate soft materials without the limitations present in other cantilevers.
"Whether we are considering biological systems or other complex, self-assembling nanostructures, this organization will be done in a liquid," says Paul Ashby, a Molecular Foundry staff scientist who led this research in the Foundry's Imaging and Manipulation of Nanostructures Facility. "If we have an investigative probe that excels in this environment, we could image individual proteins as they function on the cell surface."
Says Babak Sanii, a post-doctoral researcher in the Foundry, "Shrinking the cantilever down to nanoscale dimensions dramatically reduces the force it applies, but to monitor the movements of such a small cantilever, we needed a new detection scheme."
Rather than measuring the cantilever's deflection by bouncing a laser off it, Ashby and Sanii place the nanowire cantilever in the focus of a laser beam and detect the resulting light pattern, pinpointing the nanowire's position with high resolution. The duo say this work provides a launching pad for building a nanowire-based atomic force microscopes that could be used to study biological cells and model cellular components such as vesicles or bilayers. In particular, Ashby and Sanii hope to learn more about integrins, proteins found on the surface of cells that mediate adhesion and are part of signaling pathways linked to cell growth and migration.
"No present technique probes the assembly and dynamics of protein complexes in the cell membrane," adds Ashby. "A dynamic probe is the holy grail of soft matter imaging, and would help determine how protein complexes associate and disassociate." |
"High sensitivity deflection detection of nanowires," by Babak Sanii and Paul D. Ashby, appears in Physical Review Letters and is available in Physical Review Letters online. ###
This work at the Molecular Foundry was supported by the DOE's Office of Science.
The Molecular Foundry is one of the five DOE Nanoscale Science Research Centers (NSRCs), premier national user facilities for interdisciplinary research at the nanoscale. Together the NSRCs comprise a suite of complementary facilities that provide researchers with state-of-the-art capabilities to fabricate, process, characterize and model nanoscale materials, and constitute the largest infrastructure investment of the National Nanotechnology Initiative. The NSRCs are located at DOE's Argonne, Brookhaven, Lawrence Berkeley, Oak Ridge and Sandia and Los Alamos National Laboratories. For more information about the DOE NSRCs, please visit
nano.energy.gov.
Berkeley Lab is a U.S. Department of Energy national laboratory located in Berkeley, California. It conducts unclassified scientific research and is managed by the University of California. Visit our website at
www.lbl.gov.
Contact: Aditi Risbud
asrisbud@lbl.gov 510-486-4861
DOE/Lawrence Berkeley National Laboratory
















1 comment:
Laser activated cantilevers should probe further into nano/picoscale structural detail for bioscience modeling, and that all depends on the data density of the atomic topological function used. Recent advancements in quantum science have produced the picoyoctometric, 3D, interactive video atomic model imaging function, in terms of chronons and spacons for exact, quantized, relativistic animation. This format returns clear numerical data for a full spectrum of variables. The atom's RQT (relative quantum topological) data point imaging function is built by combination of the relativistic Einstein-Lorenz transform functions for time, mass, and energy with the workon quantized electromagnetic wave equations for frequency and wavelength.
The atom labeled psi (Z) pulsates at the frequency {Nhu=e/h} by cycles of {e=m(c^2)} transformation of nuclear surface mass to forcons with joule values, followed by nuclear force absorption. This radiation process is limited only by spacetime boundaries of {Gravity-Time}, where gravity is the force binding space to psi, forming the GT integral atomic wavefunction. The expression is defined as the series expansion differential of nuclear output rates with quantum symmetry numbers assigned along the progression to give topology to the solutions.
Next, the correlation function for the manifold of internal heat capacity energy particle 3D functions is extracted by rearranging the total internal momentum function to the photon gain rule and integrating it for GT limits. This produces a series of 26 topological waveparticle functions of the five classes; {+Positron, Workon, Thermon, -Electromagneton, Magnemedon}, each the 3D data image of a type of energy intermedon of the 5/2 kT J internal energy cloud, accounting for all of them.
Those 26 energy data values intersect the sizes of the fundamental physical constants: h, h-bar, delta, nuclear magneton, beta magneton, k (series). They quantize atomic dynamics by acting as fulcrum particles. The result is the exact picoyoctometric, 3D, interactive video atomic model data point imaging function, responsive to software application keyboard input of virtual photon gain events by relativistic, quantized shifts of electron, force, and energy field states and positions. This system also gives a new equation for the magnetic flux variable B, which appears as a waveparticle of changeable frequency. Molecular modeling and chip design engineering application software developer features for programming flow are built-in.
Images of the h-bar magnetic energy waveparticle of ~175 picoyoctometers are available online at http://www.symmecon.com with the complete RQT atomic modeling manual titled The Crystalon Door, copyright TXu1-266-788. TCD conforms to the unopposed motion of disclosure in U.S. District (NM) Court of 04/02/2001 titled The Solution to the Equation of Schrodinger.
Post a Comment